Bacterial Protocols
Contents
Preparation of DH10BAC competent cells
To
baculovirus resources
Stock solutions
LB Medium: Autoclave
1O g tryptone, 5 g yeast extract, and 5 g NaCl in 1 L.
Antibiotic solutions: To
baculovirus resources
CaCl2 solution: 60mM
CaCl2 15%(v/v) glycerol, 10mM PIPES, pH adjusted to
7.0 with KOH. Filter sterilize.
Protocol
- Inoculate a single colony of E.coli DH10BAC into
5mls of LB medium containing tetracycline at 10 µg/ml
and kanamycin at 50 µg/ml. Grow overnight at 37°C
with shaking (250 rpm).
- Inoculate 4 mls of the culture into 400ml LB medium containing
tetracycline at 10 µg/ml and kanamycin at 50 µg/ml
in a 2 litre flask. Grow at 37°C with shaking (250rpm) to
an OD590 of 0.375. (This procedure requires that
cells be growing rapidly ie. early or mid log phase).
- Aliquot culture into 8 x 50ml pre-chilled, sterile polypropylene
tubes and leave the tubes on ice 5 to 10mins. Centrifuge cells
at 4000xg, 4°C for 15 mins.
- Resuspend (gently) each pellet in 10ml ice cold CaCl2
solution. Centrifuge 15 mins at 4000xg, 4°C.
- Resuspend each pellet in 10mls cold CaCl2 solution.
Keep resuspended cells on ice for 30mins. Centrifuge 15 mins
at 4000xg, 4°C.
- Resuspend each pellet completely in 2mls of ice cold CaCl2
solution. Aliquot 250 µl into prechilled, sterile polypropylene
tubes. Freeze immediately at –70°C.
Preparation of transposition plates
To
baculovirus resources
Stock solutions
Aqueous solutions of compounds should be sterilized by filtration
through a 0.22 µm filter. Do not filter sterilize compounds
dissolved in organic solvent. Store in small aliquots in light
tight containers at -20°C. Magnesium ions are antagonists
of tetracycline. Use media without magnesium salts for selection
of bacteria resistant to tetracycline.
| Stock Solution |
Concentration (x1000) |
| Kanamycin |
50 mg/ml in water |
| Tetracycline |
10 mg/ml in ethanol |
| Gentamycin |
7 mg/ml in water |
| Bluo-gal |
100 mg/ml in DMSO |
| IPTG |
40 mg/ml in water |
Plates
Autoclave L-Agar. Cool to 55°C and then add stock solutions.
| Stock Solution |
Final concentration in agar |
| Kanamycin |
50 µg/ml |
| Tetracycline |
10 µg/ml |
| Gentamycin |
7 µg/ml |
| Bluo-gal |
100 µg/ml |
| IPTG |
40 µg/ml |
Mix the agar solution before pouring plates under sterile conditions.
Plates should be stored in the dark at 4°C, and are stable
for up to four weeks.
Transposition protocol
To
baculovirus resources
SOC Medium:
2% Bacto tryptone, 0.5% Bacto yeast extract, 10mM NaCL, 2.5mM
KCl, 10mM MgCl2, 10mM MgSO4, 20mM glucose,
filter sterilized.
- Thaw the DH10BAC competent cells on ice.
- Dispense 100 µl of the cells into 1.5 ml microfuge tubes.
- Add 1 µg recombinant donor plasmid (in 5 µl) and gently mix
the DNA with the cells.
- Incubate the mixture on ice for 30 mins.
- Heat shock the mixture by transferring to 42°C water bath
for 45 secs.
- Chill the mixture on ice for 2 mins.
- Add 900 µl SOC medium to the mixture, and transfer culture
to a universal.
- Place the universal in a shaking incubator at 37°C with
medium agitation (225 rpm) for 4-6 hours. The longer incubation
period generates more white recombinant clones.
- Serially dilute the cells, using SOC medium, to 10-1,
10-2 (i.e. 100 µl of transposition mix: 900 µl of
SOC medium = 10-1 dilution, use this to further dilute
10-fold to give 10-2 dilution). Spin down 800 µl
cells, discard 700 µl and resuspend in remaining 100 µl.
- Place 100 µl of each dilution (resuspended, neat, 10-1,
10-2) on the plates and spread evenly over the surface.
- Incubate for 48 hours at 37°C (colonies are very small
and blue colonies may not be discernible prior to 24 hours).
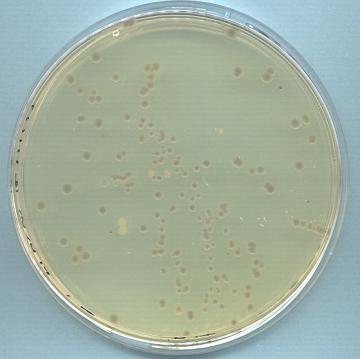 |
Recombinant white and non recombinant blue colonies
of E.coli DH10Bac. Competent DH10Bac cells were
transformed with a recombinant pFastbac construct, plated
out onto Bluogal containing media and incubated for
48 hours at 37°C. Cleavage of Bluogal by functional
ß-galactosidase in non recombinants resulted in
blue colonies. Transposition, however, caused insertional
inactivation of the bacmid encoded alpha peptide gene,
resulting in white colonies. |
Isolation of recombinant bacmid DNA
White colonies contain the recombinant bacmid, and therefore,
are selected for isolation of recombinant bacmid DNA. Before
isolating DNA, candidate colonies are streaked to ensure they
are truly white.
- Pick 4 white colonies and streak to fresh plates to verify
the phenotype. Incubate for 48 hours at 37°C.
- From a single colony confirmed as having a white phenotype
on plates containing Bluo-gal and IPTG, innoculate a 4 ml LB
medium supplemented with 50 µg/ml kanamycin, 7 µg/ml gentamicin,
and 10 µg/ml tetracycline. Grow at 37°C to stationary phase
(up to 24 hours) shaking at 250 to 300 rpm.
- Combine 0.9 ml recombinant bacmid culture with 0.1 ml sterile
glycerol, and store at -80°C.
- Transfer 1.5 ml of culture to a 1.5 ml microcentrifuge tube
and centrifuge at 14,000xg for 1 min.
- Remove the supernatant and resuspend each pellet in 0.3 ml
of solution 1 (15 mM Tris HCL pH 8.0, 10 mM EDTA, 100 µg/ml
RNase A). Add 0.3 ml of solution II (0.2 N NaOH, 1% SDS) and
gently mix. Incubate at room temperature for 5 mins.
- Slowly add 0.3 ml of 3 M potassium acetate pH 5.5, mixing
gently during addition. A thick white precipitate of protein
and E.coli genomic DNA will form. Place the sample on ice for
5 to 10 mins.
- Centrifuge for 10 mins at 14,000xg. During the centrifugation,
label another microcentrifuge tube and add 0.8 ml isopropanol
to it. Gently transfer the supernatant to the tube containing
isopropanol. Avoid any white precipitate material. Mix by gently
inverting tube a few times and place on ice for 5 to 10 mins.
At this stage, the sample can be store at -20°C overnight.
Centrifuge the sample for 15 mins at 14,000 x g at room temperature.
- Remove the supernatant and add 0.5 ml 70% ethanol to each
tube. Centrifuge for 5 mins at 14,000 xg at room temperature.
- Remove as much of the supernatant as possible.
- Air dry the pellet briefly, 5 to 10 mins, at room temperature
and dissolve the DNA in 40 µl sterile water. Store at
-20°C.
Confirmation of transposition by PCR
To
baculovirus resources
We routinely use the Expand High Fidelity
PCR System cat. No. 1732650 from Roche Diagnostics.
| Bacmid |
1 µl |
| 10x Expand HF buffer with 15mM MgCl |
5 µl |
| 2.5 mM dNTP mix (dGTP, dTTP, dATP, dCTP)* |
4 µl |
| 15 pmol/µl M13/pUC forward primer |
1 µl |
| 15 pmol/µl M13/pUC reverse primer |
1 µl |
| Expand enzyme |
0.75 µl |
| Water |
37.25 µl |
|
Total=
|
50 µl |
*To prepare 2.5 mM dNTP mix, combine equal quantities of
10 mM dGTP, 10 mM dTTP, 10 mM dATP and 10 mM dCTP.
- Set up PCR as above
- Place tube in PCR machine with heated lid.
- If the expected product is less than 5 kb, use the following
cycling conditions: (93°C x 3 mins) x 1, (94°C x 45 secs, 55°C
x 45 secs, 72°C x 5 mins) x 35. For larger products, add 1 minute
extension time per kb of DNA.
- Electrophorese 5 µl PCR on 1% agarose gel in electrophoresis
buffer, and stain with ethidium bromide and visualize DNA under
U.V. Light.
The expected results from the PCR are as follows:
| Sample |
PCR Product (approx
size) |
| Bacmid alone |
300 bp |
| Bacmid transposed with pFastBac 1 |
2,300 bp |
Bacmid transposed with pFastBac HT
|
2,430 bp |
| Bacmid transposed with pFastBac DUAL |
2,562 bp |
Insertion of your gene into any of the pFastBac donor plasmids
will result in an increase in the size of the PCR product.
This increase in PCR product corresponds to the size of your
gene of interest.
